Structural databases contain data objects including trees, graphs, or a set of interrelated labeled points in two, three, or higher dimensional space.
Example data objects:
- protein secondary and tertiary structure,
- phylogenetic trees,
- neuroanatomical networks,
- parse trees,
- molecular diagrams,
- XML documents.
This demo shows a search engine capable of answering the following two types of queries on structural database D containing graphs:
-
[nearest-neighbor query]
- Given a query object o, which items o' in D are closest to o?
-
[discovery query]
- Which substructures approximately occur in the items in D?
Some screen shots illustrating the discovery query:
1. A database of four chemical compounds (graphs).
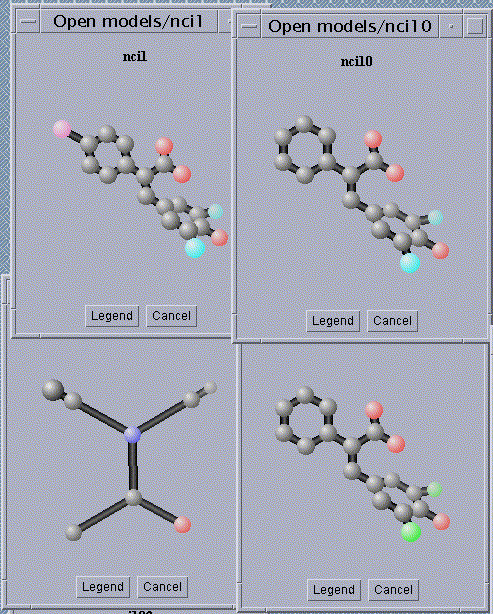
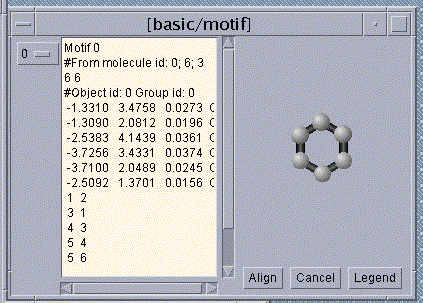
2. A substructure, found by our algorithm, that approximately occurs
in the chemical compounds in the database.
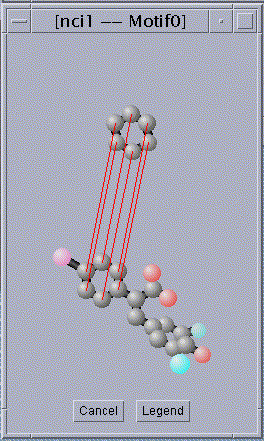
3. An alignment between the found substructure and
one of the chemical compounds in the database.
Underlying techniques:
- A technique based on topological relationships (heuristics for subgraph isomorphism).
- A technique based on physical position (geometric hashing).
- Heuristic related to simulated annealing.
- Algorithm runs in time proportional to the number of edges, though the constant of proportionality depends on the length of time spent doing the simulated annealing.
Related URLs:
http://web.njit.edu/~wangj/sigmod.html