MirID Software Manual
MirID is a tool for predicting whether an RNA sequence is a pre-miRNA or not. This tool is developed for prediction the pre-miRNA sequences of
twenty one speices: Arabidopsis thaliana, Caenorhabditis briggsae, Caenorhabditis elegans, Canis familiaris, Ciona intestinalis, Danio rerio, Drosophila melanogaster, Drosophila pseudoobscura
Epstein barr virus, Gallus gallus, Homo sapiens, Macaca mulatta, Medicago truncatula, Mus musculus, Oryza sativa, Physcomitrella patens, Populus trichocarpa
Pristionchus pacificus, Rattus norvegicus, Schmidtea mediterranea, and Taeniopygia guttata, based on the algorithms of Feature Mining and Ada Boost.
The algorithm is described in Ling Zhong, Jason T. L. Wang, Dongrong Wen, Virginie Aris, Patricia Soteropoulos, and Bruce A. Shapiro.
"Effective Classification of MicroRNA Precursors Using Combinatorial Feature Mining and Boosting Methods".Paper link.
MirID
1. Download this package from the download link above.
2. tar and unzip the package using
tar -zxf MirID.tar.gz
3. Change directory to the main directory
cd MirID
4. Download and install packages ViennaRNA and LIBSVM under the directory MirID.
Note: All the codes inside the package are executable based on your linux/unix or Cygwin system has installed perl and C/C++.
perl predict.pl <species code> <input file>
Species code can be found below in the Species Code Table. Here are examples for an input file and the output after running the above command line.
Input Example
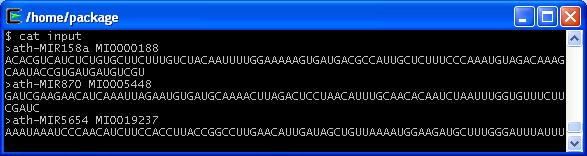 Output Example
Output Example
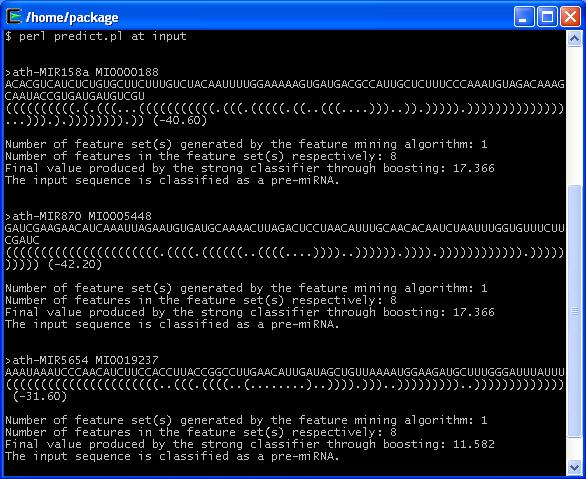
| Species
|
Code
|
| Arabidopsis thaliana
|
at
|
| Caenorhabditis briggsae
|
cb
|
| Caenorhabditis elegans
|
ce
|
| Canis familiaris
|
cf
|
| Ciona intestinalis
|
ci
|
| Danio rerio
|
dr
|
| Drosophila melanogaster
|
dm
|
| Drosophila pseudoobscura
|
dp
|
| Epstein barr virus
|
eb
|
| Gallus gallus
|
gg
|
| Homo sapiens
|
hs
|
| Macaca mulatta
|
mm
|
| Medicago truncatula
|
mt
|
| Mus musculus
|
mu
|
| Oryza sativa
|
os
|
| Physcomitrella patens
|
pp
|
| Populus trichocarpa
|
pt
|
| Pristionchus pacificus
|
pr
|
| Rattus norvegicus
|
rn
|
| Schmidtea mediterranea
|
sm
|
| Taeniopygia guttata
|
tg
|
The MirID core code has been written by Ling Zhong.
The RNA sequence folding tool is from Vienna RNA.
The SVM tool is from LIBSVM.
Please contact Ling Zhong for any further questions, comments, or suggestions.
last update: Mar 01 2017