Research Summary
Rhythms can be found nearly everywhere in biology and are fundamental to brain function. My research program seeks to uncover mechanisms underlying biological timekeeping, neuronal rhythm generation, and the disruption of rhythmicity associated with certain pathological conditions. In my work I use data-driven mathematical modeling, numerical simulation, and dynamical systems analysis to study biological systems from the subcellular scale up to networks of thousands of neurons. My interests in biological oscillations span a wide range of time scales and application areas, including daily (circadian) rhythms such as the sleep/wake cycle, the respiratory rhythm with several breaths per minute, and the alternations in visual perception that occur every few seconds during binocular rivalry.
- Research Interests: Dynamical Systems, Mathematical Neuroscience, Multiscale Modeling, Data Assimilation, Circadian Rhythms, Central Pattern Generators, Deep Learning
Research Projects
-
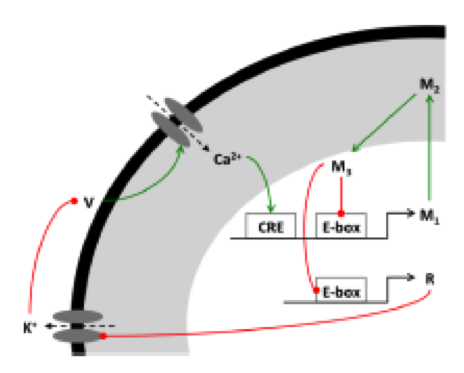
Circadian (~24-hour) rhythms offer one of the clearest examples of the interplay between different levels of organization within the brain, with rhythmic gene expression leading to daily rhythms in neural activity, physiology and behavior. The main output signal of the master circadian clock in mammals has long been believed to be a simple day/night difference in the firing rate of neurons within the suprachiasmatic nucleus (SCN). My work in collaboration with Hugh Piggins (University of Manchester) challenges this theory, and demonstrates that a substantial portion of SCN neurons exhibit a more complex and counterintuitive set of electrical state transitions throughout the day/night cycle.
One of my current research goals is to provide a mathematical understanding of the subcellular, cellular, and network properties underlying these daily transitions in SCN electrical state and the functional roles they play in the mammalian circadian clock. SCN neurons contain intracellular molecular clocks composed of gene regulatory feedback loops. The conventional wisdom is that all SCN neurons communicate their timing signal to other parts of the brain through higher firing rates during the day than at night. This view is countered by the Piggins lab's recent identification of two classes of SCN neurons with distinct firing patterns. One class of neurons did not fire action potentials (APs) at all in the middle of the afternoon---the time of day when SCN neurons were thought to be firing their fastest.
Despite the ability of individual SCN neurons to produce oscillations autonomously, reliable rhythmicity and synchronization require intercellular signaling. How intercellular signaling synchronizes and modulates the precision of cellular rhythms remains unclear. The complexity of the circadian timekeeping system calls for the use of mathematical modeling and theory to aid our understanding of pacemaking across the SCN network.
Existing mathematical models of the SCN network incorporate detailed intracellular molecular clock mechanisms, but only very simple electrical dynamics (or none at all) and phenomenological modeling of the coupling between cells. The models I am developing employ biophysical descriptions of SCN neuron membrane dynamics and synaptic connections, and significantly advance the state of the art by integrating slow genetic oscillator models (time scale of hours) and fast neural excitability dynamics (time scale of milliseconds). These models are designed to elucidate the relationship between the molecular clock and SCN neural activity, a major open question in the circadian field.
In a new collaboration with Ravi Allada (Northwestern University), we have demonstrated that NCA localization factor 1 links the molecular clock to sodium leak channel activity. Through a combination of modeling and experiments, we have found that antiphase cycles in sodium and potassium leak conductances drive membrane potential rhythms in master pacemaker neurons in both mammals and the fruit fly Drosophila. Thus, this "bicycle" mechanism is an evolutionarily ancient strategy for the neural mechanisms that govern daily sleep and wake.
-
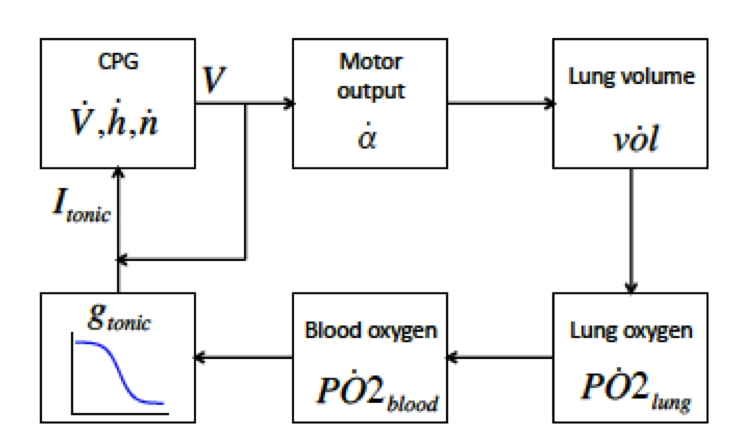
Incorporation of sensory feedback is essential to guide the timing of rhythmic motor processes. How sensory information influences the dynamics of a central pattern generating circuit varies from system to system, and general principles for understanding this aspect of rhythmic motor control are lacking. It has been realized for sometime, however, that the mechanism underlying rhythm generation in a central circuit when considered in isolation may be different from the mechanism underlying rhythmicity in the intact organism.
To explore this issue, we have developed a closed-loop model of respiratory control incorporating a conductance-based central pattern generator (CPG), filtering of CPG output by the respiratory musculature, gas exchange in the lung, metabolic oxygen demand, and chemosensation. The model exhibits the coexistence of two dynamically stable rhythmic behaviors corresponding to eupnea (normal breathing) and tachypnea (excessively rapid breathing). The latter state represents a novel failure mode within a respiratory control model. An artificially imposed bout of hypoxia can cause the system to leave the basin of attraction for the eupneic-like state and enter the basin of attraction for the tachypneic-like state.
We are investigating the relationship between rhythms in the intact (closed-loop) and isolated CPG (open-loop) systems and find that oscillations appear to arise from two distinct mechanisms. We also find that conductances endogenous to the Butera-Rinzel-Smith model of the respiratory CPG. can lead to spontaneous autoresuscitation after short or mild bouts of hypoxia.
This work is in collaboration with Peter Thomas (Case Western Reserve University) and Christopher Wilson (Loma Linda University).
-
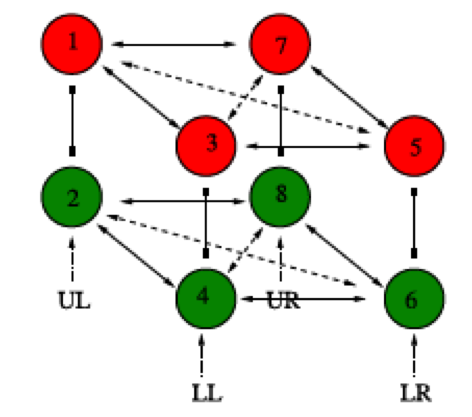
Binocular rivalry is the alternation in visual perception that can occur when the two eyes are presented with different images. For example, if one eye is shown an image of a monkey and the other eye a jungle scene, rather than perceiving a superimposition of the monkey and the jungle, the subject will typically perceive the monkey image for a while, then the jungle image, back to the monkey, and so on. Binocular rivalry experiments offer a tool for exploring neural processes involved in visual awareness and perceptual organization. Hugh Wilson has proposed a class of neuronal network models that generalized rivalry to multiple competing patterns. The networks are assumed to have learned certain patterns, and rivalry is identified with time periodic states that have periods of dominance of different patterns. My collaborators and I analyzed the structure of Wilson networks, and have shown that in certain cases the high-dimensional models can be reduced to a low-dimensional system where the dynamics are well understood. In particular, we proved that the rivalry dynamics are partially organized by a symmetry-breaking Takens-Bogdanov bifurcation, and obtained new results on paths through the unfolding of this type of singularity.
In recent work, we have formulated Wilson networks to make predictions that are directly testable via simple psychophysics experiments. For instance, Kovács et al. reported a surprising finding that has since attracted much attention: when subjects were presented two scrambled images consisting partially of a monkey and partially of a jungle scene, they perceived alternations between unscrambled images of monkey only and jungle only. We constructed a simple Wilson network to model this experiment and found that in addition to the learned patterns (the scrambled images), the network also supports "derived" patterns corresponding to the unscrambled images. Thus, the Wilson framework and equivariant bifurcation theory make completely natural this otherwise surprising experimental result.
This work is in collaboration with Marty Golubtisky (Ohio State University).
-
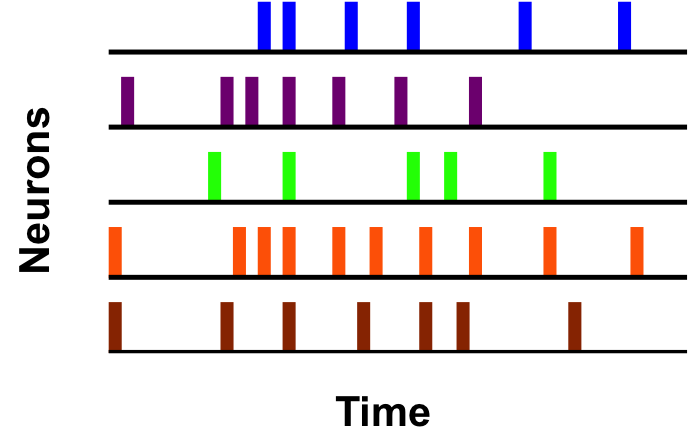
In recent years, multielectrode arrays (MEAs) have become a standard tool in neuroscience allowing researchers to record the electrical activity of multiple neurons simultaneously. However, there has not been analogous development of statistical methods to extract the temporal and spatial information from the resulting large datasets quickly and efficiently. To address this gap, I have developed techniques to discover statistically significant patterns in MEA data. These patterns are indicative of functional connections in the underlying neural tissue, and can be used to infer the graph structure of neuronal networks. Furthermore, our approach incorporates a hypothesis testing framework designed to avoid systematic prediction errors common to other network inference procedures.
Our methods are based on computationally efficient temporal data mining algorithms. This makes them well suited to meet the needs of neuroscience research going forward, as improved MEAs will enable the collection of even larger datasets. I also plan to use these methods to analyze data from SCN neurons, in order to relate neuronal firing patterns on the millisecond time scale to circadian rhythms on the 24-hour time scale.
